Dr. Michael Roehrl from Memorial Sloan Kettering Cancer Center describes the research paper he co-authored that was published by Oncotarget on February 25, 2020, entitled, “Prolyl 4-hydroxylase alpha 1 protein expression risk-stratifies early stage colorectal cancer.”
Behind the Study is a series of transcribed videos from researchers elaborating on their oncology-focused studies published by Oncotarget. Visit the Oncotarget YouTube channel for more insights from outstanding authors.
—
My name is Michael Roehrl, I’m a physician scientist at Memorial Sloan Kettering Cancer Center in New York city, faculty member in the department of pathology, and also a faculty member in the human oncology and pathogenesis program. Today, I will talk about a recent publication from my laboratory on colon cancer, specifically the discovery of new protein biomarker that characterizes early stage colon cancer.
Colon cancer is a very prevalent disease, affecting millions of people around the world. The incidence of colon cancer has lately they’ve been increasing specifically affecting younger and younger patients. So there’s an urgent need to understand the disease better, and also to identify markers, biomarkers that predict whether or not a particular cancer is aggressive or not. The focus of our publications specifically is on what we call early stage colon cancer. These are cancers that are still relatively small and not very deeply invasive into the colon.
The study of early stage colon cancer is particularly important because while surgery is the primary approach to treat these cancers, there’s often a question whether or not the patient should also receive chemotherapy, or other targeted therapies either before or after surgery, to try to prevent recurrence for later regrowth or metastasis of the tumor. So focus on early stage colon on cancer is particularly important these days, and so we focused on a very large cohort of patients with very good clinical follow up and identified using an integrated proteomic approach that uses latest generation mass spectrometry of proteins to identify a particular protein called prolyl 4-hydroxylase alpha-1 as a marker that when it’s highly upregulated predicts poor outcome, specifically in patients with early stage colon cancer.
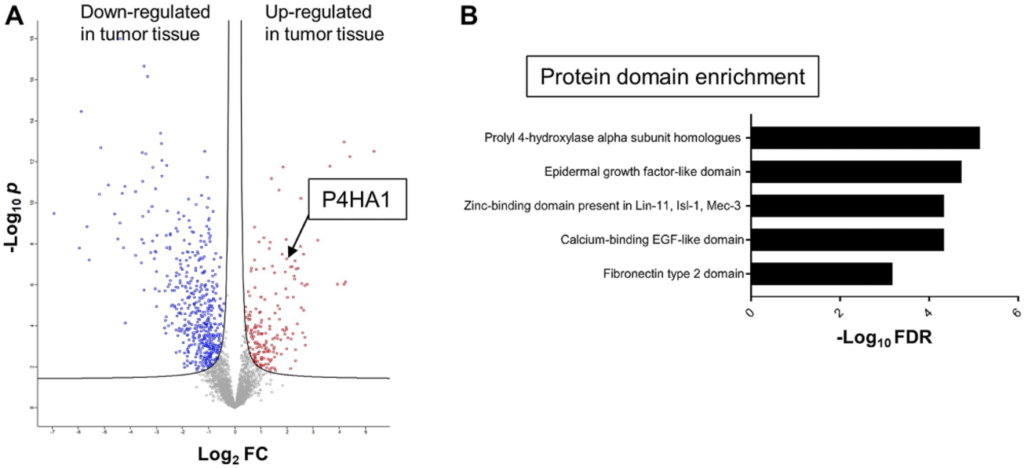
Now, why is this important? As I said, in introduction, early stage colon cancer, something we diagnose more and more commonly, but it is also one of those diseases or a stage of disease that often poses a clinical therapeutic conundrum: Should we treat the patient more aggressively or less aggressively? Do we have a chance of watch and wait, or do we need to put the patient on chemotherapy right after surgery?
So this has been a long and ongoing debate for early stage colon cancer and our study delivers a new and very promising protein biomarker that hopes to resolve, or at least contribute, to a better understanding of this conundrum. I will also highlight that the approach that we took, which is protein-centered, doing deep proteomics by mass spectrometry is something that is really cutting-edge and becoming more and more important in cancer research. Much of traditional cancer research still focuses largely on genome sequencing and transcriptional assays of messenger RNA abundance in cancers. While this is extremely useful, it really doesn’t interrogate the machines of lives, the proteins that carry out the chemistry in the cancer directly.
So looking at proteins quantitatively by biophysical tools like high sensitivity, high resolution mass spectrometry, is something that is extremely promising for the future of cancer research overall. So as I said, we took a unique approach in that we studied first a cohort of frozen tissues, frozen tumors from patients who had undergone surgery using this latest approach of mass spectrometry based proteomics. Based on that, we then hold in on a number of proteins and pathways that appear to be differentially upregulated. The really cool part was that we had a second large cohort of more than 700 patients who had been followed for many years clinically in terms of their outcomes and treatment histories, et cetera, was used then to validate and independently test the set of proteins that we identified by mass spectrometry proteomics in frozen tissues, and we used a fewer tools like immunohistochemistry, western blotting and other types of analyses in this very large cohort of over 700 patients.
What I found really cool was, first of all, that the quantitative mass spectrometer we used on a smaller discovery cohort was fully validated in the second set of independent patients that was much, much larger. But it also really allowed us to very specifically pinpoint the prognostic power of this new enzyme prolyl 4-hydroxylase, and that we were able to interrogate whether or not, for example, microsatellite status of colon cancer – which is a very common molecular test that’s performed – actually highlighted that so-called microsatellite stable colon cancer is more sensitive to this new biomarker in terms of predicting outcome. That is extremely important because, first of all, microsatellite stable colon cancer is the more common type, two thirds or more of patients are in the microsatellite stable category. Secondly, the microsatellite stable categories, the one for which there really aren’t any good current therapeutic options that are developing in a microsatellite unstable cases, for example, immuno-oncology and immunotherapy, has proven somewhat effective, but this does not directly apply to microsatellite stable patients.
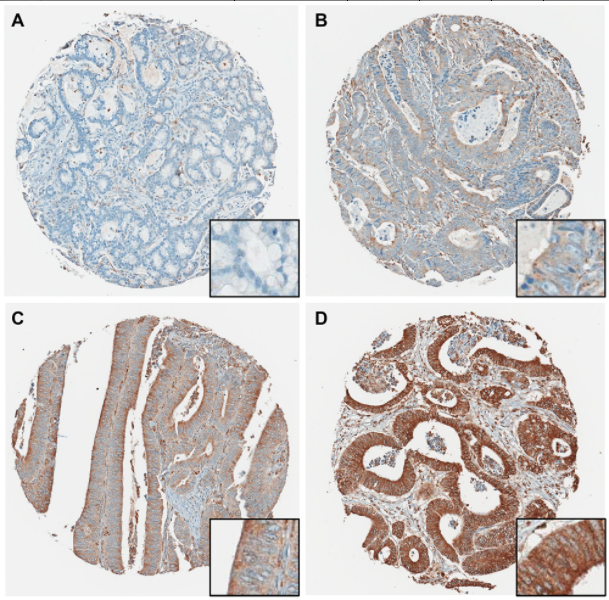
So having biomarkers that stratified risk specifically for early stage patients within the microsattelite stable cohort is particularly powerful. I found that very intriguing and really exciting to see that come out of our research. So the paper that we published on Oncotarget really is a significant first step for our laboratory to develop what I described as prognostic and predictive biomarkers in cancer overall, specifically in colon cancer.
So we’re now expanding our work to larger patient cohorts, looking prospectively at patients, how they do, and really also bring the technology of mass spectrometry proteomics into the clinic as a diagnostic tool that will be used for patient care, patient risk stratification, and also for making sure patients receive the correct treatment. So in addition to the specific finding of this one particular biomarker in the paper, I think it’s also, for us, a path way forward, a road forward to developing mass spectrometry proteomics into a key discipline for the diagnostics in cancer.
Over the next few years, I’m sure we will be seeing a number of additional protein biomarkers, pathway related biomarkers, that are very functional. And again, I would like to contrast it to the current state or status quo, which are, is very nucleic acid focused, mutation focused, genome based or transcription based. All these things are extremely useful, but we shouldn’t forget that pretty much all the drugs we have today, whether it’s small molecules, antibodies, CAR T-cells, all these targeted therapies in cancer really interact directly with proteins and not nucleic acid. So this study of biomarker discovery and study of biomarkers that are protein based in cancers, colon cancer being a very important example here, is going to grow.
I think for us, at least in my laboratory, this paper and a number of papers that we’ve published since are really very important steps towards achieving the goal of what we call proteome based diagnostics and proteome based therapy guidance in cancer. I would like to thank our funding agencies, the National Cancer Institute, and also Memorial Sloan Kettering Cancer Center grant through the Cycle for Survival Equinox Innovation Award. These have been instrumental for us to form these experiments and move forward in cancer research. Specifically I want to thank the first author of the paper, Atsushi Tanaka, an extremely talented postdoctoral fellow in my laboratory, a trained pathologist and an excellent protein mass spectrometrist, who has been really driving this project to success.
Click here to read the full study published by Oncotarget.
YOU MAY ALSO LIKE: More Oncotarget Videos on LabTube
—
Oncotarget is a unique platform designed to house scientific studies in a journal format that is available for anyone to read without a paywall making access more difficult. This means information that has the potential to benefit our societies from the inside out can be shared with friends, neighbors, colleagues, and other researchers, far and wide.
For media inquiries, please contact media@impactjournals.com.

