Dr. Herbert Levine and Dr. Mohit Kumar Jolly describe their research that was published by Oncotarget on July 7, 2020, entitled, “Epigenetic feedback and stochastic partitioning during cell division can drive resistance to EMT.”
Behind the Study is a series of transcribed videos from researchers elaborating on their oncology-focused studies published by Oncotarget. A new Behind the Study is released each Monday. Visit the Oncotarget YouTube channel for more insights from outstanding authors.
—
Dr. Herbert Levine
Hi, my name’s Herbert Levine. I’m a faculty member in physics and bioengineering at Northeastern University, and I’m the director of the Boston branch of a National Science Foundation Center, which devotes energy to applying ideas from physics and mathematics to problems in the living world, including biology, including, in this particular case, some aspects of cancer progression and cancer therapy.
The paper which I want to talk about, at the most general level, is a continuing part of our effort to understand the process called EMT. EMT stands for epithelial mesenchymal transition, or sometimes transformation. It’s a process that was originally discovered when it was noticed that some cells spontaneously transformed from being sedentary epithelial cells, all attached together, forming epithelial tissues, to cells that were more mesenchymal, which meant they were more motile, they left their original tissue, migrated throughout the developing body to other regions, and then transformed themselves into final products once they got to their destination.
It turned out that this process was not only operative in developmental systems, but also seemed to play a major role in cancer metastasis. In cancer, the initial start of cancer is when typically epithelial cells … most cancers arise from epithelial tissues. Typically, those cells begin to grow uncontrollably. For whatever reason, usually some type of genetic abnormality, cells lose their self control, they start dividing, and they start forming a tumor mass in some part of the body. We label different types of cancers by which part of the body that initial tumor takes place: breast, pancreas, liver, etc.
Initial tumors, as long as they stay in one part of the body, are not particularly good for you, but we have a variety of therapies which are quite effective, for example, surgery, for example, localized radiation therapy. It’s only when cancer metastasizes, which is when it spreads throughout the body, that you run the risk of actually dying from the disease, because then you are exposed to the possibility of multiple organ failure throughout the body, and no therapy that can then counteract that. In order to get from their primary tumor to the rest of the body, cells that initially were epithelial as part of the cancer need to become motile. They need to start being able to move, and one of the ways they can do this is by undergoing this EMT transition.
Because metastasis is the primary cause of death in most tumors, there’s been a huge emphasis in the last, let’s say, decade or two in the cancer community on understanding what are the underlying molecular processes that enable cells to undergo this transition. And of course, the eventual hope is to develop therapies, which would cause this to stop.
The path to understanding this, of course, goes from looking at experiments in laboratories, to experiments in animals, such as mice, and eventually to studying the process in humans. So if one goes back and looks at the experiment in laboratories, one thing that’s been noticed is that there are signals which can induce this transformation in cells, so that gives us a clue as to what could be happening in patients. But we’ve also observed a great deal of heterogeneity in the different cells, that when you take a series of cells that look like they came from the same type of cell and apply these signals to them, rather than getting uniform response, you get a wide variety of responses. This has been enabled by new technology in the biology world, where one can measure details of single-cell behavior rather than cell population behavior.
One of the things that was noticed in this context of EMT was that, if you applied an EMT signal to a population of cells, you would notice that a significant proportion of the cells looked like they were resistant to those signals. That you would apply what you would think would be a typical dose that would induce this transition, but nonetheless, those cells, because of some uniqueness in their molecular makeup, or their molecular dynamics, somehow were being resistant to that.
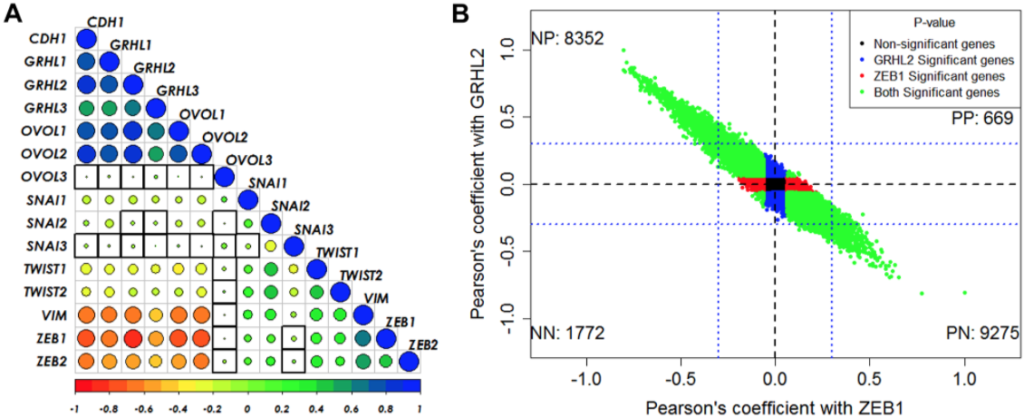
This paper attempted to address, by means of mathematical, physical modeling tied to experimental data analysis, what could be possible causes of this mechanism. What could be possible mechanisms that underlie this very interesting, and somewhat surprising, feature that’s been seen experimentally in EMT.
Mohit, maybe you should explain what some of the possibilities were, and how we began to investigate that.
Dr. Mohit Kumar Jolly
Hi, my name is Mohit Kumar Jolly. I’m an assistant professor at Center for Biosystem Science and Engineering at Indian Institute of Science in Bangalore. I’ll follow up on what “Herbie” talked about in this paper.
We started looking into these processes of EMT when I was a graduate student with “Herbie” a couple years ago. They’re trying to ask questions about how is the dynamics of this process looking like, because, at that point of time, our experimental cancer biology colleagues mostly had readings only at day zero and day end, when this process ends. There was very little understanding of how this process exactly happens, and that took us by surprise, given the importance of this process, not only in metastasis, but also in the years, over the past years, as we realized in terms of evading, say, even the immune system, various different drugs out there, changing the metabolism, et cetera.
So EMT sort of happens to act as a central fulcrum through which cells seem to adapt to various different bottlenecks that they face during the process of metastasis. When we had developed the mathematical models for it, our predictions were that EMT is not as blindly a process as black and white, as all or none, kind of a process as people were automatically thinking about at that point of time. So we made these predictions that there can be multiple stable states in which cells can maintain themselves for long durations of time, and over the past couple of years, those predictions have been experimentally tested, validated, such as what we call hybrid cells have been seen in the clinic, as well. More importantly, it has been shown that, not only these hybrid cells in exist, but the existence of these hybrid cells is actually of paramount importance for the process of metastasis.
If you deplete these hybrid cells, particularly, then metastasis can decrease significantly, is what some of the experimental studies have shown. So now, we wanted to ask the question, if a cell is becoming mesenchymal and then it is supposed to revert back to being epithelial once it reaches a distant organ, what you need in cells is their ability to come back, the diversability aspect of this entire process. So we asked this question, that is, are these processes always reversible? One of the previous mathematical models that we have developed for this suggested that that’s not necessarily always the case. There comes a tipping point in between after which it is possible that cells do not come back, even when you withdraw the signal for prolonged durations of time. Our enthusiasm was increased by the fact that there was experimental evidence, both collected by our collaborators and others existing in the published literature, which actually showed this is possible.
Now, in this particular paper published in Oncotarget, we wanted to take this question further and ask that is EMT, or when cells come back from mesenchymal, is that always reversible, or in other words, as Herbert was mentioning, does every cell always undergo the transition? So if you have a population of cells, can you say that every single cell will undergo a transition at some point of time, or there will always be cells which will just resist any change in their behavior? So here we looked at two specific mechanisms. One of them relates to the epigenetic feedback, which comes back to some of the chromatin’s remodeling that has been shown experimentally to be happening during this particular process, and the other is the process of stochastic noise asymmetry in distribution of molecules when a cell divides.
I’ll explain these processes one by one. What has been reported, to not only EMT, but in various other similar processes where cells transition from one state to one other, that there are mechanisms at the chromatin level, regulatory processes, which somehow lock the cell in the new state. That is important because you need the other state to be stable as well. You don’t need it to be completely meta stable, that it can be pushed out. So there are these mechanisms which happen, which somehow change the regulatory network underlying and enabling this more or less irreversible kind of transition.
We developed a simple mathematical model to represent the dynamics and ask this question, is this our process, is this our mechanism through which some cells can continue to resist undergoing EMT throughout? And we observed, indeed, that was the case.
We wanted to look at also another process, which is the asymmetry in cell division. When a cell divides, for instance, in the EMT field, we still do not completely understand that when a cell is undergoing EMT, the same cell, which is becoming from E to M, or is the cell dividing and giving rise to two daughter cells, one of which stays epithelial and the other becomes mesenchymal. The timescales at which these processes are studied, at least in the laboratory systems over a week or so, are timescales at which cells are continuously dividing, so we wanted to see what exactly is the effect, at a cell population, that might be happening due to cell division.
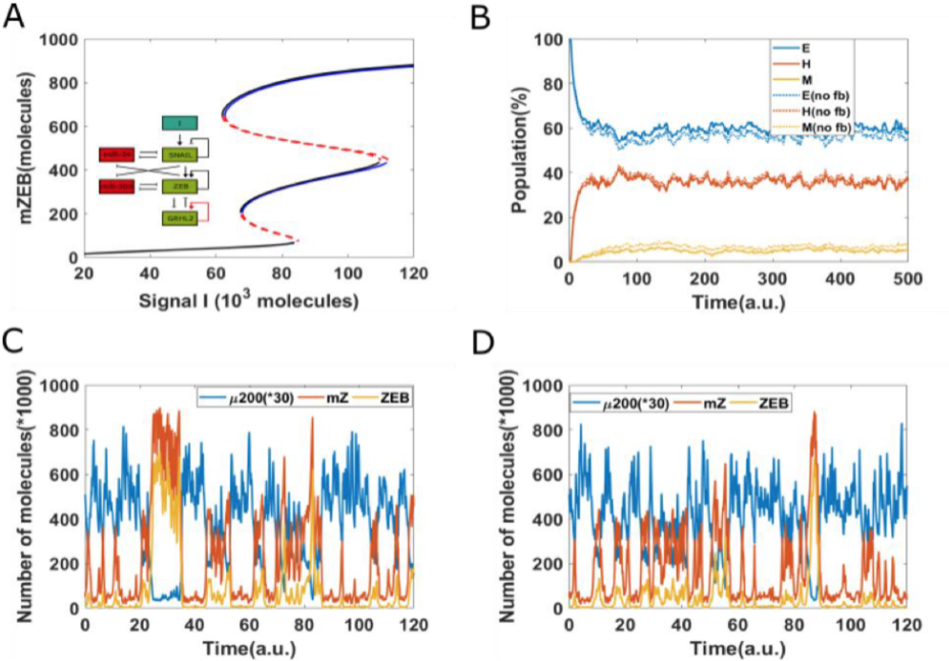
Now, every time a cell divides, there can be some differences in the partitioning of molecules. You cannot expect both the rotor cells to get the exact number of molecules for all different biochemical species in a cell which was there in the parent molecule. There is always going to be some noise. There’s always going to be some asymmetry.
So we wanted to ask this question that, given there is some bare minimum level of asymmetry, can this be yet another mechanism through which some cells can resist undergoing this transition? Because when there is this asymmetry, then some cells are likely to be more resistant, versus some cells are likely to be less resistant. So, even if you start from a homogeneous population, you are actually creating more and more heterogeneity, and you’re creating more and more subpopulations, some of which are more likely, as compared to the parent, and some of which are less likely, as compared to the parent, to undergo these transitions.
So these are the two of the processes that we have identified, which can contribute to this. Fortunately, there were recent studies at a single cell level, also, which showed that, in a cell population, this kind of phenomena are indeed seen, and they did show at least correlative evidence that epigenetic players can be associated with that. There is no cause and effect that particular study has shown so far. So what our approach establishes is the potentially causatory link that those epigenetic changes are not just correlated, but they may be actually underlying this completely different responses of the two or more subpopulations actually in the process.
Dr. Herbert Levine
I wanted to say a little bit more about the general problem of trying to incorporate epigenetic effects into these regulatory networks. By regulatory networks, we’re usually thinking back to when most of the work on those networks was being done in relatively simpler organisms, bacteria, yeast, et cetera. And in those cases, essentially, most of what’s interesting and happening dynamically to cells can be described by the interaction of transcription factors, proteins that determine DNA expression, and those are responsible for changing the cell state from one part to the other.
The interaction of transcription factors, thought of as a network of interacting chemical interactions, has been a very fruitful way of creating models that can explain and predict data in those systems. But when we get to humans, but more generally, when we get to complex animals, it turns out that just thinking about these networks as independent transcription factors that are operating in an absolute context really doesn’t seem to be the right paradigm for the field, because the field has learned over the last 10 years that there are large scale changes in chromatin structure, in accessibility of genes, in how genes affect each other, which sort of sets a context for how these transcription factor networks are going to behave.
We’ve identified various specialized transcription factors. They’re sometimes called pioneers, which somehow set the stage for all the other transcription factors in the networks. The field has been trying to understand how to incorporate these new insights into a next generation set of models. Our approach has been to take very simplified versions of that to just get started, to see if even simplified versions can begin to give us insights into what’s going on. Other people have taken more, let’s say, detailed modeling perspective, where they’ve tried to incorporate extremely large number of details about how these DNA reorganization, chromatin chain processes, actually work, and of course, the problem with that is then you’re subject to not knowing all the parameters, not knowing which is relevant in which case, not knowing all the couplings, not even knowing all the players, initially.
The field is still working very hard on trying to move forward with this, how epigenetics really affects cell transformations, how it affects memory of what cells you were when you go through division. And this is a paper which, in the general sense, contributes to that effort, even though, of course, its focus is on a specific transformation on the EMT process.
Also I wanted to mention in this regard that the aspect of the EMP process which we focused on, which is the role of a specific molecule called GRHL2, is not arbitrary. It’s partially because GRHL2 is known to play a vital role in maintaining the chromatin structure relevant for epithelial-like states. So GRHL2, if it’s enabled to be activated, can very easily suppress EMT. That’s what we were looking for, and, therefore, we focused on mechanisms that would control, in an epigenetic way, the ability of GRHL2 to accomplish that.
![Figure 6: Reversibility of EMT starting from epithelial state (miR-200 = 17,000, mZEB = 50, ZEB = 10,000 molecules), a cell is treated by different time duration (5, 10, 20 arbitrary units [a.u], as marked by arrow) of high EMT-inducing signal (I = 125,000 molecules), corresponding to the {H, M} bistable region.](https://www.impactjournals.com/wp-content/uploads/2021/09/Screen-Shot-2021-09-23-at-1.57.13-PM.png)
We have in mind a more general question, but of course, this particular paper takes that general question, constructs model relevant for a specific process, and then tries to see to what extent it can explain data regarding EMT.
Dr. Mohit Kumar Jolly
Follow up on that, I think one of the overarching themes that this paper connects to is also growing literature on what one can call non-genetic heterogeneity or phenotypical heterogeneity in the context of cancer. So again, the molecular oncology field has been largely dominated by thinking in terms of mutations and genetic changes, which are irreversible, are heritable, and continue forever because they’re encoded in the DNA. It’s hard wired. What some of our previous work, and extended by this particular work, we have are highlighting in mechanisms through which this heterogeneity can exist, even when genetic background is completely identical, or what is called an isogenic cells, in that case.
So be it the ability of cells to exist in multiple states, be it their ability to lock certain decisions or unlock certain decisions based on the chromatin rewiring, be it the noise or asymmetry in division of molecules, these are happening largely independently of the genetic background. The genetic background, of course, may be altering the rates of one versus the other. And genetic and non-genetic are again, perhaps, not as clearly distinguishable as we are putting it now, but nonetheless, we want to highlight that there are other mechanisms which have been studied in plenty in simpler micro organisms, such as bacteria and yeast. There is literature on phenotypical heterogeneity or non-genetic heterogeneity for a long period of time, but somehow the cancer community has not really caught up on that, so this is one more contribution at conceptual or a thematic level that this paper tries to make.
Dr. Herbert Levine
I wanted to add to that. One of the directions for our future work is the role of this type of phenotypic heterogeneity, specifically in the context of drug resistance, because what’s been observed in many different types of cancers is a drug that initially starts out creating a relatively good response, shrinking the tumor, reducing the tumor load, the symptoms, nonetheless fails relatively quickly as the tumor rebounds, starts to grow again. Of course, eventually tumors can become really resistant to cancer therapy drugs, usually by genetic mutations, but there’s an intermediate timescale, perhaps over the course of weeks or months, when there hasn’t been really enough time for a mutation to arise that specifically can account for why the drug is failing, but nonetheless, the drug begins to fail.
What’s believed to be the case is that the intrinsic heterogeneity in the tumor, this phenotypic heterogeneity, creates a variety of responses, creates a subpopulation of cells, that are not really sensitive to the particular drug. And, of course, if you kill other cells that are competing with them for resources, metabolic resources, then the more tolerant subpopulation will gain ascendancy, and will take over the tumor population, and your drug will stop to have a significant effect.
Then, if you wait long enough, this tolerant population, which is still maybe growing at a slow rate, or maybe just sort of static, will eventually evolve a genetic change, which will then completely defeat the drug, making the drug no longer useful whatsoever. So it’s now believed that, in many of these processes, the pathway and root to drug resistance is phenotypic heterogeneity. So studying phenotypic heterogeneity in a variety of contexts will offer insight into what can be done to combat it in the case of drug resistance, and that’s actually one of the future directions that we are looking at collaboratively with a number of cancer biology research groups.
Click here to read the full study published by Oncotarget.
YOU MAY ALSO LIKE: More Oncotarget Videos on LabTube
—
Oncotarget is a unique platform designed to house scientific studies in a journal format that is available for anyone to read—without a paywall making access more difficult. This means information that has the potential to benefit our societies from the inside out can be shared with friends, neighbors, colleagues, and other researchers, far and wide.
For media inquiries, please contact media@impactjournals.com.
